Molecular Mapping and Marker Assisted Selection of Soybean Mosaic Virus Resistance Gene RSC12 in Soybean 
 Author
Author  Correspondence author
Correspondence author
Legume Genomics and Genetics, 2010, Vol. 1, No. 8 doi: 10.5376/lgg.2010.01.0008
Received: 07 May, 2010 Accepted: 03 Jul., 2010 Published: 08 Dec., 2010
Ma et al., 2010, Molecular Mapping and Marker Assisted Selection of Soybean Mosaic Virus Resistance Gene RSC12 in Soybean, Legume Genomics and Genetics Vol.1 No.8 (DOI: 10.5376/lgg.2010.01.0008)
The P1, P2, F1 plants, F2 population and F2:3 lines from the cross of Qihuang22×Nannong1138-2 were inoculated with the soybean mosaic virus (SMV) strain SC12 for identification of their resistance in the greenhouse. Qihuang22 and F1 individuals were resistant(R), and Nannong1138-2 were susceptible(S). The F2 population segregated in a ratio of 3(R):1(S), and the F2:3 lines exhibited a segregation pattern of 1(R):2(Segregating):1(S). These results indicated that a single dominant gene controlled the resistance to SC12. A F2 population of Qihuang22×Nannong 1138-2 with 219 individuals was constructed for molecular mapping of resistance gene RSC12 to soybean mosaic virus in soybean. Linkage analysis with bulk segregant analysis (BSA) demonstrated that the resistance gene RSC12 was located on the linkage group F and linked with seven SSR markers. The order and genetic distance of markers linked with RSC12 were Sat_297 6.4 cM Sat_234 4.9 cM Sat_154 1.1 cM Satt114 0.7 cM SOYHSP176 1.6 cM Satt334 2.4 cM RSC12 6.3cM Sct_033. The marker-assisted selection (MAS) efficiency of SSR markers Satt334 and Sct_033 was evaluated in F2, F3 and F4 populations. The results showed that the MAS efficiency of Satt334 and Sct_033 was more than 85%, and that the MAS efficiency reached as high as 95% when these two markers were co-used. Therefore, the two SSR markers can be used effectively in selecting for resistance genes RSC12 instead of inoculation identification.
Soybean mosaic virus (SMV) disease is one of the most destructive viral diseases in soybean (Glycine max (L.) Merr.) production worldwide, which resulted in substantial yield losses and seed-quality deterioration. Planting resistance varieties is the most economical, effective and environmentally friendly approach for controlling the disease. However, traditional phenol- typic selection for varieties resistant to SMV is time- consuming and easily restricted by inoculation and identification conditions, which might be unavailable for many breeding agencies. Molecular marker-assisted selection (MAS) has been proved to be a highly efficient breeding approach to select resistant lines. The co-dominant simple sequence repeat (SSR) marker, which has many advantages such as abundant poly- morphism, excellent repeatability, simple and rapid testing and so on, has been widely used as a tool in MAS breeding programs.
So far, there were some SSR markers identified for MAS of soybean cyst nematode (SCN) resistance, such as Satt309 (Cregan et al., 1999; Wang et al., 2003), Sat_168 (Cregan et al., 1999), Satt038 (Mudge et al., 1997; Prabhu et al, 1999), Satt130 (Mudge et al., 1997), Sat_162 (Meng et al., 2003), Satt610 (Meng et al, 2003) and so on. However, the researches on soybean resis- tance to SMV were mainly focused on mapping of the resistance genes to SMV. Yu et al. (1994), Jeong et al. (2002) and Hayes et al. (2000) found the molecular markers closely linked to SMV resistance genes, Rsv1, Rsv3 and Rsv4, and located the three resistance genes on linkage groups F, D1b and B2, respectively. Zhang et al. (1998, Chinese Science Bulletin, 43(20): 2197- 2202) and Wang et al. (2004) mapped the genes sep- arately controlling resistance to SMV strain Sa, SC8, SC9, N1, N3. Zheng et al. (2001) mapped the resistance gene to the SMV3 strain in northeast of China by using random amplified polymorphic DNA (RAPD). Researches reported above mostly adopted molecular markers such as restriction fragment length polymer- phism (RFLP) and RAPD which presented low avai- lability to map the resistance gene. Recently,Li et al. (2006) and Bai et al. (2009) located the SC14, SC11 resistance genes on linkage group F, and found the SSR markers closely linked to the resistance genes, respecttively. But there hasn’t reports on molecular markerassisted selection for SMV resistance genes yet.
In this paper, we determined the inheritance mode of resistance to strain SC12 in Qihuang22, identified molecular markers linked to the resistance gene, loc-ated the resistance gene on the soybean genetic linkage map, and verified MAS efficiency of the markers closely linked to the resistance gene in order to provide theory and method guidance for the resis- tance breeding programs to SMV and resistance genes pyramiding.
1 Results
1.1 Inheritance of resistance to SMV strain SC12 in soybean
The inoculation reaction results showed that Qihuang22 and F1 from the cross Qihuang22×Nannong1138-2 displayed resistance to the SMV strain SC12, while Nannong1138-2 was susceptible. F2 segregation population exhibited a good fit to the expected ratio of 3 resistant (R): 1 susceptible (S), and F2:3 lines segregated with an acceptable fitness to 1 R: 2 seg- regating : 1 S (Table 1). These results indicated that a single dominant gene, designated as RSC12, controlled resistance to the strain SC12 in Qihuang22.
![]()
Table 1 Segregation analysis of reaction to SMV strain SC12 in Qihuang22×Nannong1138-2
1.2 Gene mapping of RSC12 with SSR markers
7 SSR markers, Sat_297, Sat_234, Sat_154, SOYH- SP176, Satt114, Satt334 (Figure 1) and Sct_033, which presented polymorphic between Qihuang22 and Nannon1138-2 as well as the resistant and susceptible bulks, were identified based on the bulk segregant analysis (BSA). The 7 markers showed a good fitness to 1:2:1 ratio in the F2 segregation pop- ulation by χ2 tests (Table 2). Furthermore, analysis of linkage by using software MAPMAKER/EXP 3.0b indicated that the 7 markers were all linked to the resistance gene RSC12 And RSC12 was located on linkage group F based on the soybean integrated linkage map by Song et al. (2004). The order and genetic distance between the markers and RSC12 were Sat_297 6.4 cM Sat_234 4.9 cM Sat_154 1.1 cM Satt114 0.7 cM SOYHSP176 1.6 cM Satt334 2.4 cM RSC12 6.3cM Sct_033 (Figure 2).
|
Figure 1 Analysis for genotype of SSR marker Satt334 closely linked to RSC12 in F2 population of Qihuang×Nannong1138-2 |
|
Table 2 Segregation analyses for molecular markers closely linked to RSC12 in a F2 population from the cross of Qihuang22 and Nannong1138-2 |
.png) Figure 2 The genetic linkage map of SMV resistance gene RSC12 |
1.3 The MAS efficiency for the resistance gene RSC12
The phenotype of F2, F3 and F4 populations were identified by artificial inoculation, and the genotype of each plant from F2, F3 and F4 was tested by the SSR markers closely linked to RSC12, Satt334 and Sct_033. The data obtained was used to evaluate the MAS efficiency of RSC12 by Satt334 and Sct_033 so as to verify their reliability and practicability in MAS.
In the F2 population, among the 164 plants showing resistant phenotype after inoculation, 159 presented resistant genotype tested by Satt334, and 155 presented resistant genotype tested by Sct_033. The coincidental rates were 97.0% and 94.5%, respectively. Of the 55 susceptible plants identified by inoculation, 54 showed susceptible genotype by Satt334, and 49 displayed susceptible genotype by Sct_033. The coincidental rates were 98.0% and 89.1%, respectively (Table 3). 153 resistant genotype plants tested by Satt334 and Sct_033 were all resistant phenotype by inoculation, which indicated that MAS efficiency for resistant plant by Satt334 and Sct_033 was as high as 100% (Table 4).
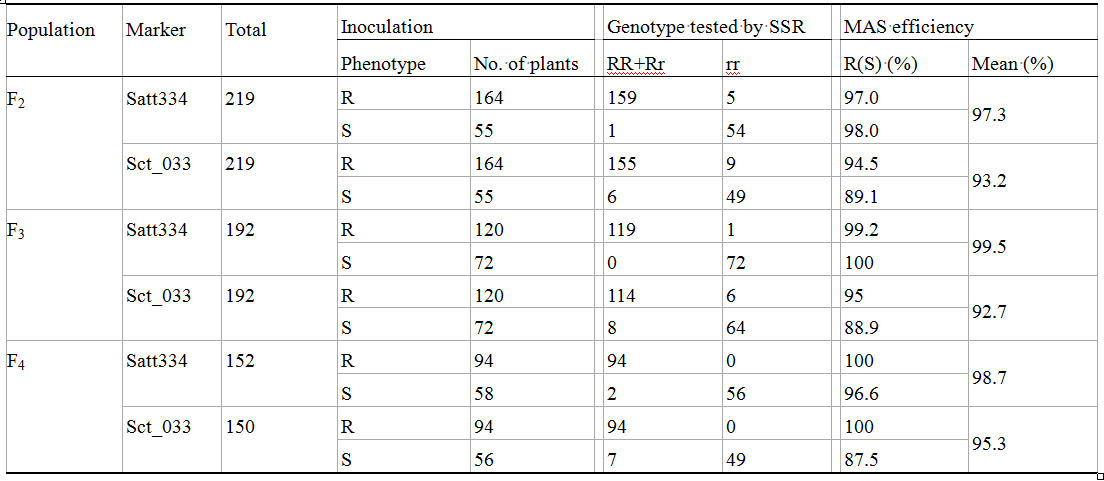 Table 3 Coincidental rate between SSR detection and inoculation in F2, F3 and F4 population |
|
Table 4 MAS efficiency when two markers Satt334-Sct_033 flanking RSC12 were co-used |
Similarly, MAS efficiency for the resistance plants in F3 and F4 populations were all more than 85% by using either Satt334 or Sct_033 (Table 3), and up to 100% by the two markers co-used (Table 4).
This result indicated that the resistance of the plants tested by the SSR markers Satt334 and Sct_033 showed high consistency to the one by conventional inoculation. Therefore, the two SSR markers can be used effectively in selecting for resistance gene RSC12 instead of inoculation identification.
2 Discussion
In present study, the resistance gene RSC12 from Qi- huang22 was located on linkage group F between the SSR markers Satt334 and Sct_033. Interestingly, RSC14 and RSC11 from Qihuang No.1 were also previ- ously mapped on linkage group F between Satt334 and Sct_033 by Li et al. (2006) and Bai et al. (2009), respectively. The resistance to SMV in Qihuang22 and Qihuang No.1 may be controlled by a same gene due to the latter as one of the parents of the former. And there also was the report that the region densely covered with R genes was closely linked to Satt334 and Sct_033 (Liu et al., 2000, Progress in Natural Science, 10(11): 1012-1017). The truth that whether the resis- tance to the 3 different SMV strains was controlled only by the same gene or by 3 different linked genes still need to be further explored.
The ideal markers used in MAS are based on PCR by considering experimental cost and technical feasibility. Zheng et al. (1997) considered that the available gen- etic distance between markers used in MAS and the target gene should be less than 5.0 cM. In the previous reports, most of the markers used to locate the resis- tance genes to SMV were RAPD and RFLP markers (Yu et al., 1994; Jeong et al., 2003; Hayes et al., 2000; Zhang et al., 1996; Zheng et al., 2001; Wang et al., 2004), which were applied limited in breeding program for their poor repeatability, complicated operation and high labor and time consumed. The co-dominant SSR markers Satt334 and Sct_033 identified in this research, closely linked to the resistance gene to SMV, can be used to screen the homozygous resistant plant in early generations by convenient experimental operation. Furthermore, the markers used in MAS showed high efficiency, and the efficiency for the resistance plant selection was more than 94% when one marker beside RSC12 was used and 100% when the two markers were co-used. Therefore, it is feasible for the two SSR markers to be used as a tool in SMV resistance breed- ing program.
In the recent years, breeders try their best to integrate several different resistance genes into a same elite variety, so as to improve resistance, broaden resistance spectrum, and prolong its service life in agricultural production. There is no chance to inoculate the same plant with different SMV strains simultaneously by conventional pathogen inoculation method, which is inconvenient to screen and identify the individual plant possessing multi-resistance from hybrid proge- nies. Fortunately, the molecular markers, especially SSR markers, closely linked to the target gene can be used in MAS to pyramid multi genes into a same variety quickly and accurately. The markers closely linked to the resistance gene to SMV identified in this research provide the basic information in pyra- miding the resistance genes in soybean.
3 Materials and Methods
3.1 materials
The cultivar Qihuang22, obtained from multiple cross of Yishuipingdinghuang, Qihuang No.1, Juxuan23 and Nongza9-3 (Xu et al., 2004), is resistant to SMV strain SC12, and is also multi resistant to some other different SMV strains (Shang et al., 1999; Li et al., 2006; Wang et al., 2003). F1, F2, F2:3, F3, F4 were all obtained from the cross of Qihuang22 (R)×Nanno- ng1138-2 (S).
The SMV strain SC12 is a prevalent low virulent strain distributed in Northern China Spring Planting Soybean Region, Middle and Lower Huang-Huai and Changjiang Valleys (Guo et al., 2005; Wang et al., 2005), and whose reaction on differential hosts was similar with No.2 strain groups in northeast classified by Lv et al.(1985).
3.2 Inoculation identification and resistance evaluation
The parents and their offspring were planted in plastic pots in an aphid-free greenhouse. All the young plants in pots were inoculated with the inoculum containing SC12 by gently rubbing the new leaves when the primary leaves unfolded, and once more when the ï¬rst trifoliate leaves expanded. The observations were taken 1-week after the ï¬rst inoculation, and then disease reactions to SC12 were evaluated at 3-days interval for 3 weeks. At the same time the plants with symptoms were marked to avoid disturbance from latency of symptoms. The pesticide were sprayed on time to avoid the cross infection by aphid.
Any plant with mosaic symptoms on leaves above those inoculated was regarded as susceptible. Then, the number of plants with different symptomatic reaction was calculated, respectively. Chi-square tests were performed to determine the goodness-of-ï¬t of observed segregation ratios in F2 and F2:3.
3.3 Preparation of the resistant bulk and susceptible bulk
Soybean genomic DNA was extracted from sampled leaves from the two parents and their generations by using cetyl trimethyl ammonium bromide (CTAB) method. The resistant bulk and the susceptible bulk were prepared by separately pooling equal amount of genomic DNA (about 1 μg) from 15 resistant and 15 susceptible plants randomly selected from the F2 population derived from Qihuang22×Nannong1138-2.
3.4 Simple sequence repeats analysis
The primers whose sequences were obtained from the Soybase website (http://129.186.26.94/ssr.html) were synthesized by Invitrogen Biotech Ltd. Co. (Shanghai, China).
When the SSR markers were used for screening and MAS, PCR ampliï¬cation was performed in a total volume of 10 μL containing 50 ng genomic DNA, 1.5 μL 10×PCR buffer, 2 mmol/L MgCl2, 0.3 μmol/L of each primer, 0.24 mmol/L dNTP, 0.6 U Taq polymerase. Each reaction mixture was covered with paraffin oil. Ampliï¬cations were carried out following these PCR cycling conditions: 94ºC for 3 min, followed by 30 cycles of 95ºC for 30 s, 55ºC for 30 s, and 72ºC for 40 s, with final incubation at 72ºC for 8 min.
Each PCR product mixed with 2 μL loading buffer was separated on 8% non-denaturing polyacrylamide gels, and then viewed by silver staining.
3.5 Linkage analysis
The linkage between SSR markers and the resistance gene was calculated under MAPMAKER/EXP ver- sion 3.0b (Lander et al., 1987) and transformed into cM according to Kosambi’s function (Kosambi, 1944). The logarithm of likelihood ratio (LOD) 3.0 was used as a criterion to test the linkage. The linkage map was drawn by a Microsoft Excel macro called MapDraw (Liu and Meng, 2003).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 30971815), the Key Technologies R&D Program of China (Grant No. 2006BA- D01A04), the State Key Basic Research and Development Plan of China (Grant No. 2006A10A111).
References
Bai L., Li H.C., Ma Y., Wang D.G., Liu N., and Zhi H.J., 2009, Inheritance and gene mapping of resistance to soybean mosaic virus strain SC-11 in soybean, Soybean Science, 28(1): 1-6
Cregan P.B., Mudge J., Fickus E.W., Danesh D., Denny R., and Young N.D., 1999, Two Simple sequence repeat markers to select for soybean cyst nematode resistance conditioned by the rhg1 locus, Theor. Appl. Genet., 99: 811-818 doi:10.1007/s001220051300
Guo D.Q., Zhi H.J., Wang Y.W., Gai J.Y., Zhou X.A., Yang C.L., Li K., and Li H.C., 2005, Identification and distribution of soybean mosaic virus strains in Middle and Northern Huang Huai Region of China, Chinese journal of oil crop science, 27(4): 64-68
Hayes A.J., Ma G., Buss G.R., and Saghai Maroof M.A., 2000, Molecular marker mapping of Rsv4, a gene conferring resistance to all known strains of soybean mosaic virus, Crop Sci., 40: 1434-1437 doi:10.2135/cropsci2000.4051434x
Jeong S.C., Kristipati S., Hayes A.J., Maughan P.J., Noffsinger S.L., Gunduz I., Buss G.R., and Saghai Maroof M.A., 2002, Genetic and sequence analysis of markers tightly linked to the soybean mosaic virus resistance gene, Rsv3, Crop Sci., 42: 265-270 doi:10.2135/cropsci2002.0265
Kosambi D.D., 1944, The estimation of map distances from recombination values, Ann. Eugen., 12: 172-175
Lander E.S., Green P., Abrahamson J., Barlow A., Daly M.J., Lincoln S.E., and Newburg L., 1987, MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimenta land natural populations, Genomics, 1: 174-181 doi:10.1016/0888-7543(87)90010-3
Li H.C., Zhi H.J., Bai L., Yang H., Ma Y., Liu N., and Wang D.G., 2006, Studies on inheritance and allelism of resistance genes to SMV strain SC-11 in soybean, 2006, Soybean Science, 25(4): 365-368
Li H.C., Zhi H.J., Gai J.Y., Guo D.Q., Wang Y.W., Li K., Bai L., and Yang H., 2006, Inheritance and gene mapping of resistance to soybean mosaic virus strain SC14 in Soybean. Journal of Integrative Plant Biology, 48(12): 1466-1472 doi:10.1111/j.1744-7909.2006.00365.x
Liu R.H., and Meng J.L., 2003, MapDraw: a microsoft Excel macro for drawing genetic linkage maps based on given genetic linkage data, Hereditas, 25(3): 317-321
Lv W.Q., Zhang M.H., Wei P.W., Xie S.Y., GUO J.Q., Jiang Y.Y., and Geng Y.C., 1985, Classification and distribution of strains of soybean mosaic virus in Northeast China, Acta Phytopathologica Sinica, 15(4): 226-229
Meng X., Liu X.Y., and Fang X.J., 2003, QTL mapping genes conferring resistance to race 4 of Soybean Cyst Nematode in soybean ZDD2315 (Glycine max (L) Merr.) based on public molecular genetic linkage map, Molecular Plant Breeding, 1(1): 6-21
Mudge J., Cregam P.B., Kenworthy J.P., Kenworthy W.J., Orf J.H., and Young N.D., 1997, Two SSR markers that flank the major soybean cyst nematode resistance locus, Crop Sci., 37: 1611-1615 doi:10.2135/cropsci1997.0011183X003700050034x
Prabhu R.R., Njiti V.N., Bell-Johnson B., Johnson J.E., Schmidt M.E., Klein J.H., and Lightfoot D.A., 1999, Selecting soybean cultivars for dual resistance to soybean cyst nematode and sudden death syndrome using two DNA markers, Crop Sci., 39: 982-987 doi:10.2135/cropsci1999.0011183X003900040005x
Shang Y.F., Zhao J.H., Yang C.L., Li C.S., Lu X.B., Xin X.Q., and Luo R.W., 1999, Classification and distribution of strains of soybean mosaic virus in Huang-Huai area of China, Acta Phytopathologica Sinica, 9(2): 115-119
Song Q.J., Marek L.F., Shoemaker R.C., Lark K.G., Concibido V.C., Delannay X., Specht J.E., and Cregan P.B., 2004, A new integrated genetic linkage map of the soybean, Theor. Appl. Genet., 109(1): 122-128 doi:10.1007/s00122-004-1602-3
Xu R., Shi C.E., Zhang L.F., Wang C.J., Nie C.Q., and Li J.H., 2004, Utilization of Qihuang 1 in soybean breeding in the Huanghuaihai region, Journal of Plant Genetic Resource, 5(2): 170-175
Wang W.H., Qiu L.J., Chang R.Z., Ma F.M., Xie H., and Lin F.Y., 2003, Characteristics of alleles at Satt309 loucs associcated with rhg1 gene resistant to SCN of Chinese soubean germplasm, Soybean Science, 22(4): 246-250
Wang X.Q., Gai J.U., Yu D.Y., and Zhi H.J., 2003, Idenfication of sources of soybeans resistant to new SMV strain groups in middle and lower Huang-Hhuai and Changjiang valleys, Soybean Science, 22(4): 241-245
Wang Y.J., Dongfang Y., Wang X.Q., Yang Y.L., Yu D.Y., Gai J.Y., Wu X.L., He C.Y., Zhang J.S., and Chen S.Y., 2004, Mapping of five genes resistant to SMV strains in soybean, Acta Genetica Sinica, 31(1): 87–90
Wang Y.W., Zhi H.J., Guo D.Q., Gai J.Y., Chen Q.S., Li K., and Li H.C., 2005, Classification and distribution of strain groups of soybean mosaic virus in Northern China spring planting soybean region, Soybean science, 24(4): 263-268
Yu Y.G., Saghai-Maroof M.A., Buss G.R., Maughan P.J., and Tolin S.A., 1994, RFLP and microsatellite mapping of a gene for soybean mosaic virus resistance, Phytopathology, 84: 60-64 doi:10.1094/Phyto-84-60
Zhan Y., Yu D.Y., Chen S.Y., and Gai J.Y., 2006, Inheritance and gene mapping of resistance to SMV strain SC-7 in soybean, Acta Agronomic Sinica, 32(6): 936-938
Zheng C.M., Chang R.Z., and Qiu L.J., 2001, Inheritance of resistance to SMV3 and identification of RAPD marker linked to the resistance gene in soybean, Scientia Agricultura Sinica, 34(1): 1-4
Zheng K.L., and Huang N., 1997, Outlook on the application of marker-assisted selection in rice improvement, Hereditas, 19(2): 40-44
. PDF(225KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Ying Ma
. Haichao Li
. Dagang Wang
. Ning Liu
. Haijian Zhi
Related articles
. Soybean
. Soybean mosaic virus
. Inheritance of resistance
. Gene mapping
. Marker assisted selection
Tools
. Email to a friend
. Post a comment


